Background
Cancer is a disastrous growth of cells in the body. In the most common types of cancer, this growth is brought on by a series of many mutations – accumulated throughout life and possibly influence by extrinsic factors.
Childhood cancers are different. In young age, cells have not usually attained many mutations yet, but rather there are individual catastrophic events that occur in undifferentiated cells during the orderly growth of the organism. These singular mutations throw developing cells of their designated path and result in malignancies. However, for most pediatric cancers, the exact histogenesis is not fully understood. Insight into how mutations affect development will ultimately elucidate new avenues for detecting (diagnosis), projecting (prognosis), and interfering (therapy) with aberrant development in childhood cancers.
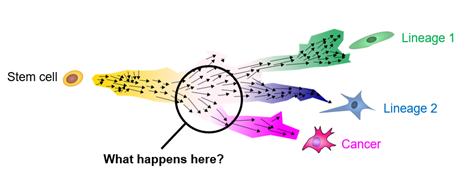
Our research
Taking apart pediatric cancer bit by bit
We are a team of computational biologists approaching childhood cancer development with a scientist’s curiosity in three facets:
Charting and modelling (cancer) cell development
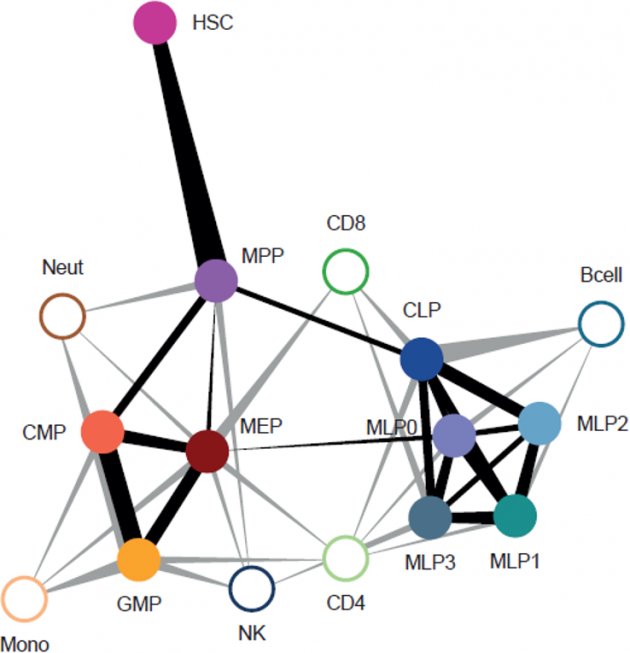
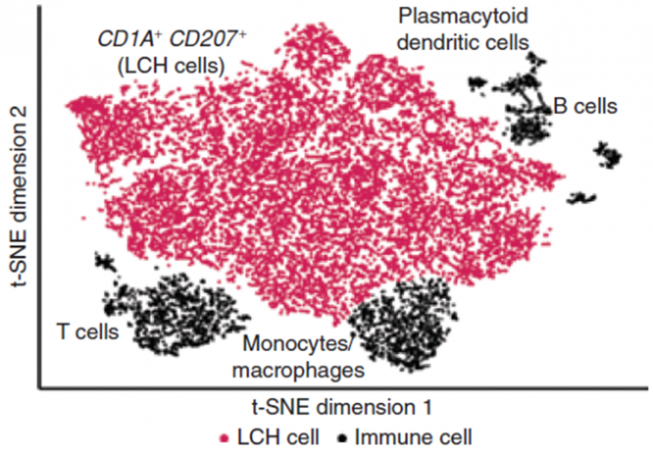
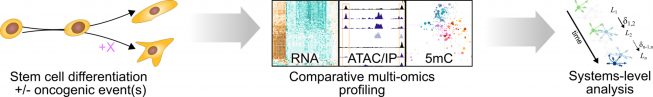
In our research, we contribute to charting the molecular changes that occur in cells during organismal development and to juxtapose those to the biology of tumors. For instance, we have profiled the transcriptome and epigenome of developing hematopoietic progenitors (see figure). Aberrant hematopoiesis is the root of the most common childhood malignancies (leukemia), but also of the rare myeloid neoplasm Langerhans cell histiocytosis (LCH). With the lab of Caroline Hutter (at CCRI), we investigated intra-tumor heterogeneity in LCH at single-cell level (see figure) and uncovered an unexpected cellular hierarchy between multiple coexisting LCH cell states. Our ongoing work focuses on embryonal tumors like Wilms tumor and neuroblastoma.
Stem-cell-based models
When do cancerous cells diverge from normal development? What are the gene-regulatory mechanisms that define these cell states and govern plasticity? To answer this sort of question, we have in recent years turned toward stem-cell-based in vitro models to study the very root of oncogenic transformation. For this purpose, we seek out and work with experts for stem cell technologies at CCRI and around the world. Together, we use the culture models as analytically tractable systems to identify the precise effects of oncogenic events on differentiating cells. Ultimately, we hope that this knowledge may help to inspire new diagnostic and therapeutic approaches in the management of childhood cancers.
High-throughput genomics and computational biology
Our research benefits heavily from technological breakthroughs in the field of functional genomics. We now routinely use high-throughput functional genomics assays (including single-cell RNA-seq, ATAC-seq, ChIP-seq, RRBS/WGBS, lineage tracing) and, more recently, spatially resolved techniques to pair cell-intrinsic states with cell-extrinsic influences. Armed with these powerful technologies, we generate rich, multi-faceted portrays of cells, which we dissect using bioinformatic pipelines combined with statistical analysis and machine learning.
Algorithms for data analysis
To advance our field, we develop algorithms for data analysis (e.g., bioinformatics workflows), we pair up with experimental research groups to test, optimize, and customize assays (e.g., we evaluated DNA methylation biomarker assays and helped to develop a massively parallel reporter assay), and we commit to open science practices by regularly sharing data, code, and resources with the community.
Links
- Lab website (more info & resources): https://cancerbits.github.io
- Twitter (social media): https://twitter.com/cancerbits
- GitHub (code): https://github.com/cancerbits
- Google Scholar (publications): https://scholar.google.at/citations?user=kvN5U0IAAAAJ
Projects and Funding

- ß-catenin Roles and Dynamics in Wilms Tumors
CCRI responsible Project Lead: Maud Plaschka
Grant from the Austrian Science Fund (FWF), ESPRIT Program
Duration: 01/09/2024 – 31/08/2027 - Oncogenic aberration of development in childhood cancer
CCRI responsible Principal Investigator: Florian Halbritter
Grant from the Austrian Science Fund (FWF), Principal Investigator Project
Duration: 01/06/2024 – 31/05/2028 - Iterative programming of blood cells (ML2Cell)
CCRI responsible Principal Investigator: Florian Halbritter
Grant from the Austrian Science Fund (FWF), TAI-1000 Ideas Program, DOI: 10.55776/TAI732
Duration: 01/01/2023 – 31/07/2024

- Tracking Ewing sarcoma origin by developmental and trans-species genomics (ORIGIN)
CCRI responsible Principal Investigators and Coordinator: Heinrich Kovar
Additional CCRI Principal Investigators: Martin Distel, Florian Halbritter
Alex´s Lemonade Stand Foundation (ALSF), Crazy 8 Initiative Award Program
Duration: 01/03/2021 to 28/02/2025
- Elucidating the role of chromosomal copy number alterations in neuroblastoma
CCRI responsible Principal Investigator: Florian Halbritter
Cancer Research UK, Discovery Programme Foundation Awards
Duration: TBD
Selected Articles
Collaborating Partners
National: Igor Adameyko (MUW), Christoph Bock (CeMM), Christa Buecker (MFPL), Matthias Farlik (MUW), Johannes Gojo (MUW), Cornelia Kasper (BOKU), Martin Leeb (MFPL), Daniela Lötsch-Gojo (MUW), Florian Grebien (VetMed), Julia Ressler (MUW). International: Stefan Barakat (Erasmus MC, NL), Keisuke Kaji (Edinburgh, UK), Minoru Takasato (JP), Anestis Tsakiridis (Sheffield, UK)
About Florian Halbritter
Florian Halbritter, PhD, has been leading the Developmental Cancer Genomics group at St. Anna Children’s Cancer Research Institute (CCRI) since 2018. He studied bioinformatics and developmental biology at the Universities of Osnabrueck (Germany) and Edinburgh (UK) and worked as a postdoctoral researcher in epigenomics and cancer biology at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences in Vienna. Halbritter’s research now focuses on the developmental origins of pediatric cancers. His team develops computational and experimental approaches to dissect the molecular and cellular foundation of tumor formation. Throughout his career, Halbritter has been a fervent supporter of collaborative and open science. He participated in multiple international consortia, contributed to open resources and methods development, and promoted genomics and computational approaches in the scientific community.
Team







