Two St. Anna CCRI projects selected for FWF’s 1000 ideas program
(Vienna, 16.8.2022) This year’s 1000 ideas program of FWF promotes another two of St. Anna CCRI’s “completely new, daring or particularly original research ideas that lie outside the current scientific understanding”. Florian Halbritter’s new approach aims to integrate machine learning directly into the experimental design and execution of manufacturing specific cell types from stem cells – with the ultimate goal to reprogram even cancer cells. Artem Kalinichenko, who was also awarded a 1000 ideas grant, is targeting abnormal tumor metabolism by identifying immunogenic cancer-specific metabolites, drugs and drug-like molecules, which could be exploited for immunotherapy.
In their project “Iterative programming of blood cells”, Florian Halbritter and Davide Seruggia, Principal Investigators at St. Anna Children’s Cancer Research Institute (St. Anna CCRI), together with their teams will optimize the process of generating blood cells in the lab. To this end, they will follow an innovative hybrid approach that merges computational and experimental design principles. “We exploit epigenomics technologies to take real time measurements of these cells as they go through their differentiation process from a stem cell towards a mature blood cell. In parallel, we closely monitor this progress by bioinformatics analysis, so that we can decide on how to adapt the differentiation conditions to approximate the target cell types,” explains Florian Halbritter.
Learning from each experimental step
According to the scientists, lab protocols to program cell identities typically involve sequential stimulations with complex cocktails of molecules. Adhering to the right sequence and timing of steps is crucial, which makes protocol design via classic screening approaches (i.e., trying out everything) challenging: As each variable is only evaluated at the very end of a long-lasting experiment, the refinement of differentiation protocols is time consuming, expensive, and only feasible for a limited set of stimulations.
To overcome these limitations and learn in real-time from each experimental step in the differentiation protocol, the team of researchers employ algorithmic principles from machine learning, and implement them directly in the design of biological experiments. “We do not simply use machine learning to interpret data generated by the experiments but instead we make the execution of the biological experiments themselves part of the algorithm,” Florian Halbritter points out. To benchmark their approach, the scientists will generate two blood cell types from undifferentiated blood progenitors. “We use next-generation sequencing to profile the chromatin status of the cells. This will tell us which condition gives the best differentiation output”, explains Davide Seruggia, whose team will generate the experimental data.
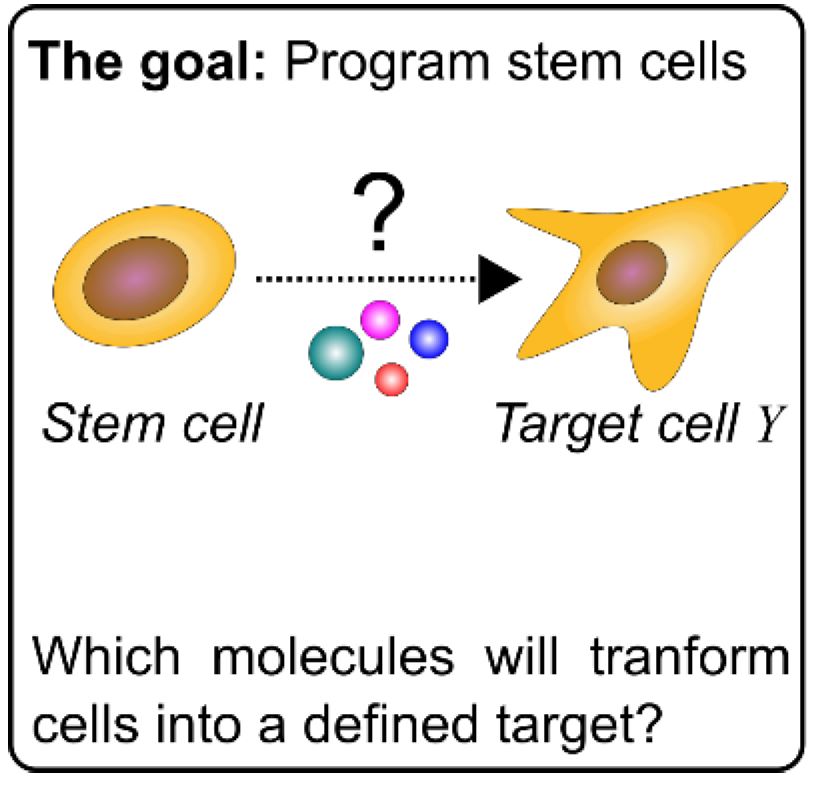
“This project is important for our work because it will be another proof that looking at chromatin and epigenetics is the best way to categorize a cell status in a cell lineage,” highlights Davide Seruggia. If successful, this new tool to reprogram cells has a potential application in regenerative medicine with the goal to replace damaged or diseased tissue, for example, to generate skin for burn victims or platelets for patients under chemotherapy. Florian Halbritter adds, “There are also potential applications in disease modeling for research purposes, but one of my ultimate goals would be to reprogram cancer cells to a less malignant state. I am hoping this will be one of the first steps in this process.”
Tumor metabolism is a universal target for the immune system
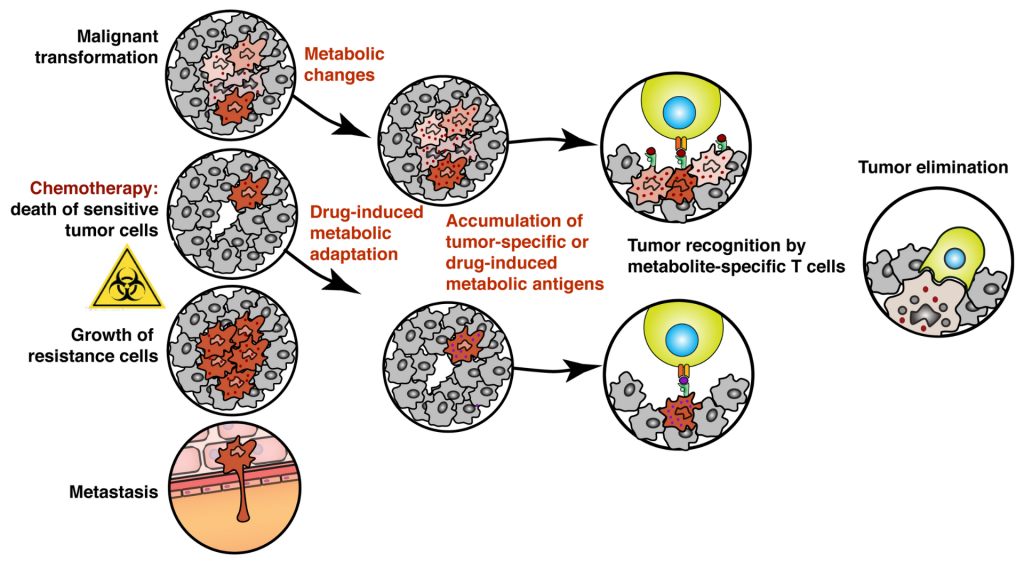
In his “Targeting Tumor Metabolism (TATUM)” project, Dr. Artem Kalinichenko investigates how to advance current treatments for different cancer types avoiding relapses and targeting drug-resistant tumors. Artem Kalinichenko, together with Univ.-Prof. Dr. Kaan Boztug, Scientific Director of St. Anna CCRI and colleagues from the Boztug lab, studies human lymphocytes sensitive to a wide range of small molecules, including those that are unique for tumor cells or accumulated in tumor cells upon treatment.
A specialized subset of immune cells in humans called cytotoxic T cells can recognize and eliminate malignant or transformed cells. Most of these T cells distinguish tumors from healthy tissues by recognizing mutated peptides presented by specialized antigen-presenting molecules called MHC I present on the surface of every cell in our body. However, most pediatric tumors do not accumulate enough mutations for their efficient recognition and elimination by the conventional, peptide-specific T cells.
To overcome this issue, the scientists will exploit the activity of metabolite-reactive so-called unconventional T cells sensitive to a wide range of organic molecules, including small metabolites, drugs, and drug-like molecules. These metabolite-specific lymphocytes are present in all individuals and scientists want to learn how to manipulate their activity to fight cancer. “We know that upon chemotherapeutic treatment tumor cells change their metabolism to adapt and survive. We hypothesize that such adaptation is forcing tumors to accumulate specific metabolites that can be recognized by metabolite-sensitive T cells. In our study, we want to find out pathways producing these molecules – we call antigens – and drugs that enhance their accumulation in tumors resulting in better immune recognition. We will then use these drugs to isolate and characterize metabolite-sensitive T cells and test their potential as immunotherapy,” explains Artem Kalinichenko.
“Targeting tumor metabolism with unconventional T cells might bring multiple opportunities and benefits, especially for pediatric oncology. Low immunogenicity of pediatric tumors to the conventional, peptide-specific T cells is one of the main problems for oncologists. Bypassing it by learning how to manipulate the activity of metabolite- and drug-specific T cells might help to improve the outcome of current treatments. The fact that many tumors regardless of their origin rely on common metabolic pathways for their survival provides us with a unique opportunity to develop a common treatment equally efficient in the different types of cancer,” highlights Kaan Boztug.
“The main innovative idea for future clinical application is to combine cellular and chemotherapeutic approaches in one. Eventually, we want to find a drug or drug combination that would have a dual action. On one hand, be highly cytotoxic and kill the cancer cells, and on the other force tumor recognition and elimination by metabolite-specific T cells. We think that this will help to overcome drug resistance and prevent metastases,” says Artem Kalinichenko.